Proteasome impairment compromises mitochondrial function in yeast
In eukaryotic cells, the proteasome is a protein complex that eliminates damaged and non-functional proteins, helping to maintain cellular homeostasis and proper functioning. Mitochondria are the center of energy metabolism and produce most energy needed for cellular functioning.
In recent years, studies have revealed that these two systems are more connected than previously thought: the proteasome participates in the quality control of proteins destined for the mitochondria, while mitochondrial metabolism influences the efficiency of protein degradation.
A new study from the Research Center for Redox Processes in Biomedicine (RIDC Redoxoma), led by Marilene Demasi at the Instituto Butantan, presents a valuable experimental model to investigate the interaction between the proteasome and mitochondrial functionality.
The research focused on the effects of proteasome dysfunction in the C76S mutant strain of the yeast Saccharomyces cerevisiae and revealed that deficiency in this system leads to increased mitochondrial oxidative stress. This effect was evidenced by elevated hydrogen peroxide (H₂O₂) release and reduced levels of peroxiredoxin 1 (Prx1), an enzyme crucial for peroxide removal. In mammals, mitochondrial Prx3 is equivalent to yeast Prx1.
“What is important in this work is that we now have a yeast strain that serves as a model to investigate the proteasome deficit in the interlocution with mitochondrial metabolism. This model didn’t exist in the literature,” highlights Demasi.
The study, conducted in collaboration with the research groups of Mario H. Barros (ICB-USP) and Luis E.S. Netto (IB-USP), both members of the RIDC Redoxoma, was published in the journal Archives of Biochemistry and Biophysics.
Redox deficit
The research used the C76S strain of Saccharomyces cerevisiae, previously characterized by Demasi’s group, in which a cysteine residue is replaced by serine in the alpha-5 subunit of the 20S proteasome. Under fermentative conditions, these cells exhibit a reduced chronological lifespan, proteasome dysfunction, and accumulation of polyubiquitinated proteins.
To explore the mitochondrial contribution in this model, the researchers first cultivated the mutant strain C76S in a respiratory medium containing glycerol and ethanol as carbon sources, aiming to stimulate mitochondrial proliferation.
In inicial phenotypic tests, they noticed that the yeast growth curve in this medium was deficient and that the cells displayed morphological alterations. These findings motivated a deeper investigation into the cells’ metabolism.
“When we evaluated the role of the proteasome, we immediately saw an accumulation of polyubiquitinated proteins, which means that the proteasome was not properly degrading proteins, since we found a deficit in the catalytic activity of the 20S proteasome, as well as its coupling to the 19S regulatory particle, which is responsible for recognizing polyubiquitinated proteins,” Demasi explained. “Next, we analyzed oxygen consumption by isolated mitochondria and were surprised to find it was very elevated. ATP production in this strain was also immense, which really caught our attention,” she added.
The ubiquitin-proteasome system plays a central role in cellular regulation. Regulatory proteins are marked with ubiquitin, a signal that allows their recognition and degradation by the proteasome when the 20S catalytic unit is coupled to the 19S regulatory unit. Oxidized or poorly structured proteins, however, can be eliminated independently of this marking by the free 20S proteasome.
Another significant finding was the increase in the activity of some respiratory chain complexes, even in the absence of changes in their abundance. Given this, the researchers decided to investigate the mitochondria’s redox status and identified an increased hydrogen peroxide release, accompanied by reduced levels of Prx 1. At the whole-cell level, they detected a change in the glutathione pool, one of the main intracellular redox buffering systems.
“In addition, we observed DNA fragmentation, an important marker of apoptosis, and the release of cytochrome c into the cytoplasm. Our conclusion was that increased oxidative stress, combined with loss of proteasome activity, drives these cells to death by apoptosis,” explained Demasi.
To test whether Prx1 plays a causal role in the observed phenotype, the team overexpressed the enzyme in the mutant strain. The intervention partially reversed the defects, clearly demonstrating that Prx1 is a key factor in the recovery of C76S cells.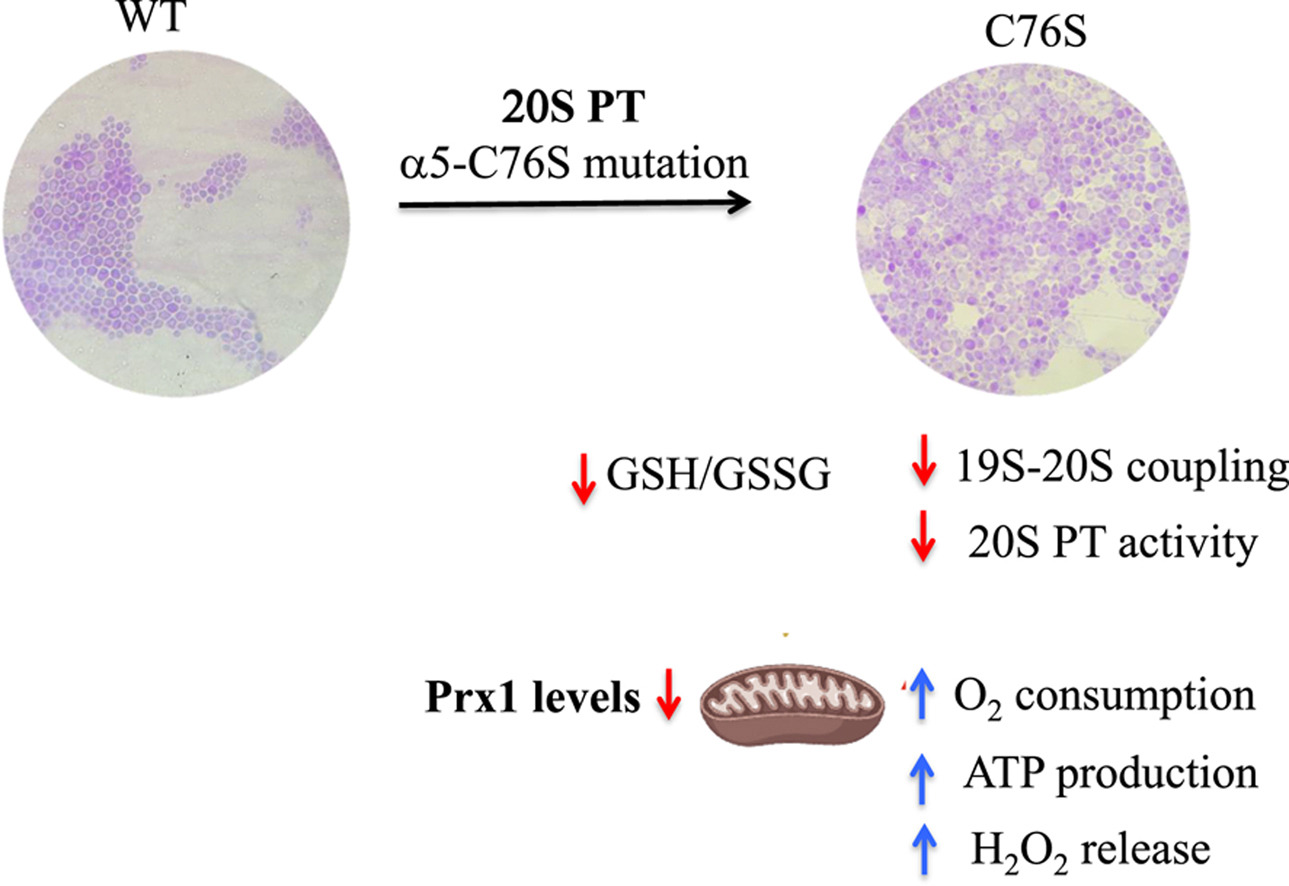
Next steps
The researchers are now working to understand why Prx1 levels are reduced in cells with compromised proteasome. “We still do not know whether there was a decrease in Prx1 gene expression, which is possible, since the proteasome also plays a role in regulating gene transcription, or whether the protein is more oxidized. It may become hyperoxidized and, therefore, be more degraded. Perhaps the excess peroxide is promoting its continuous degradation,” says the researcher.
To address these questions, the group intends to perform comparative transcriptomic and proteomic analyses between the wild-type strain and the mutant strain grown under respiratory conditions aiming to establish the mutant as a model to investigate the role of the ubiquitin-proteasome system (UPS) in cellular metabolism.
The article Decreased levels of Prx1 are associated with proteasome impairment and mitochondrial dysfunction in the yeast Saccharomyces cerevisiae by Natália Mori Avellaneda Penatti, Mário Henrique Barros, Fernando Gomes, Luis Eduardo Soares Netto, Kamila de Jesus Maciel, Vincent Louis Viala, Ana Mara Viana and Marilene Demasi, can be accessed here.
